Salvatore Milite
PhD Student @Human Technopole

PITA 02.011
Human Technopole
20157, Milan, MI
I am a graduate student in computational biology in the Lab of Prof. Andrea Sottoriva at Human Technopole, co-supervised by Prof. Giulio Caravagna at the University of Trieste. I also have a Master’s degree in Data Science and Scientifi Computing from the University of Trieste and SISSA. My main interests are statistical learning and generative models. I have years of experience in developing tools for the analysis of NGS data. Currently, I am focusing on developing deep generative methods to analyze and integrate single-cell multi-omics data, with a particular focus on understanding cancer evolution under therapy. I am also extremely excited about hierarchical models (hierachical VAEs are my obsession rn) and the theory of hierarchy in complex systems (this gives a good idea of what I am talking about)
In reality, I am basically into every challenging problem that has some maths and stats in it (bonus point if there is biology), so if you are interested in what I do or you want to talk science (and maybe have a cool scientific problem to disucss) feel free to get in touch either by email or on social media.
news
| May 25, 2024 | I will be presenting my work at the EMBO conference The many faces of cancer evolution in Rimini, see you there. |
|---|---|
| May 01, 2024 | I have been admitted to the Generative Modeling Summer School taking place in Eindhoven, June 24-28th. Very excited to learn more about my favourite topic with lots of great instructors. |
| Apr 09, 2024 | Our preprint on Multiomics Archetypal Analysis is now finally available online. |
| Jan 31, 2024 | Our preprint on exploiting Neural ODEs for computing RNA velocity in scRNA-seq in online. |
| Jan 31, 2024 | Our new work Computational validation of clonal and subclonal copy number alterations from bulk tumor sequencing using CNAqc is out on Genome Biology. |
selected publications
-
 Deep Archetypal Analysis for interpretable multi-omic data integration based on biological principlesbioRxiv, 2024
Deep Archetypal Analysis for interpretable multi-omic data integration based on biological principlesbioRxiv, 2024 -
 Clinical application of tumour-in-normal contamination assessment from whole genome sequencingNature Communications, 2024
Clinical application of tumour-in-normal contamination assessment from whole genome sequencingNature Communications, 2024 -
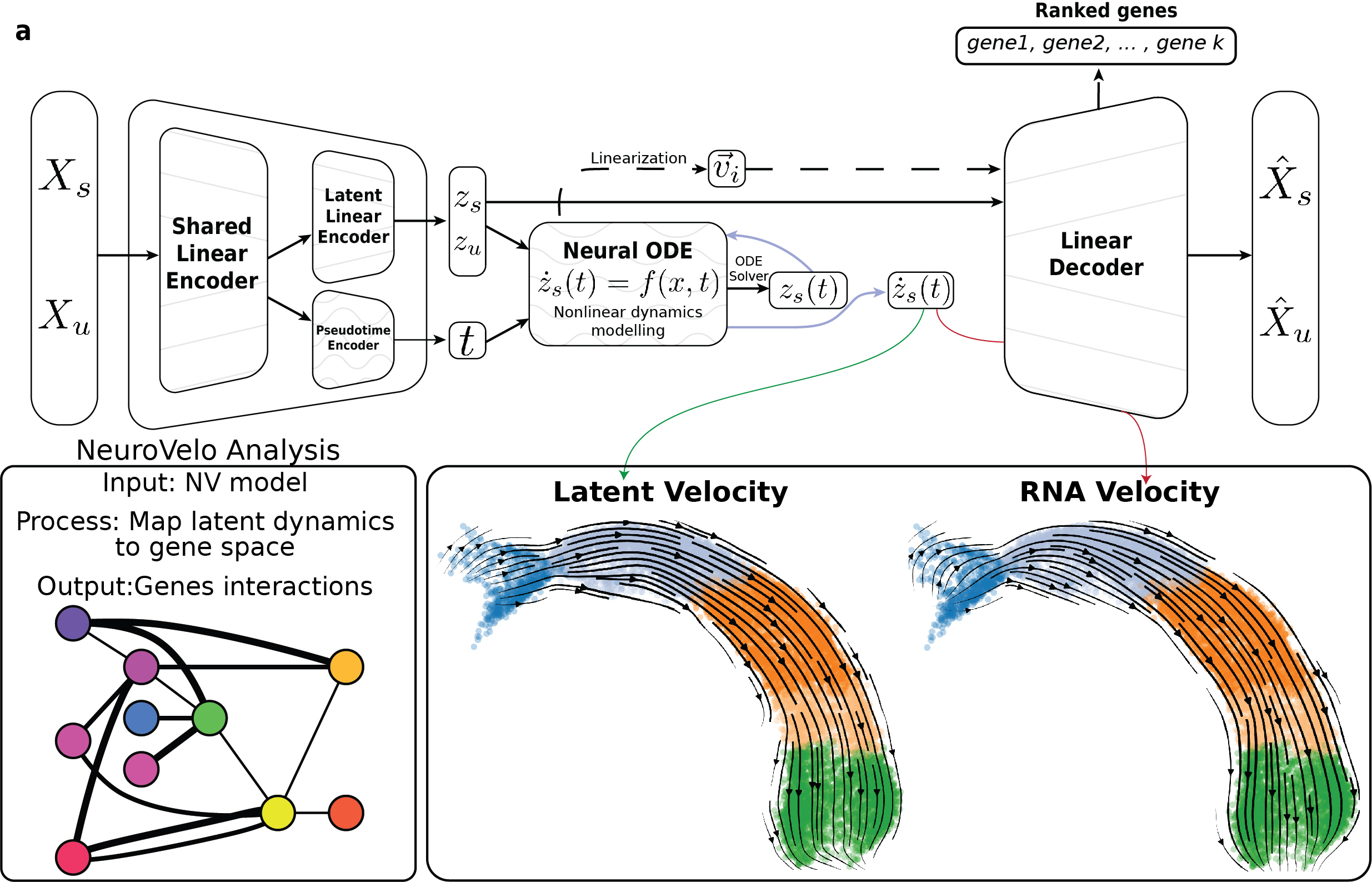 NeuroVelo: interpretable learning of cellular dynamics from single-cell transcriptomic databioRxiv, 2023
NeuroVelo: interpretable learning of cellular dynamics from single-cell transcriptomic databioRxiv, 2023 -
 Epigenetic heritability of cell plasticity drives cancer drug resistance through one-to-many genotype to phenotype mappingbioRxiv, 2023
Epigenetic heritability of cell plasticity drives cancer drug resistance through one-to-many genotype to phenotype mappingbioRxiv, 2023 -
 A Bayesian method to cluster single-cell RNA sequencing data using copy number alterationsBioinformatics, 2022
A Bayesian method to cluster single-cell RNA sequencing data using copy number alterationsBioinformatics, 2022